 Just out in the ISME journal, a paper on my favourite bacterium, Myxococcus xanthus. I did my PhD with Greg Velicer on this fascinating species which builds beautiful multicellular fruiting bodies that can be seen with the naked eye (click on my personal page underneath the page header for a publication list with pdf downloads, including a review paper). I thought it would be nice to return to the strains I isolated back then and do some population genomics. It turned out to be an interesting project although not without significant obstacles; papers are a bit like sausages, they can be very enjoyable but you do not want to necessarily know how they were made. Anyway, from the Abstract:
Just out in the ISME journal, a paper on my favourite bacterium, Myxococcus xanthus. I did my PhD with Greg Velicer on this fascinating species which builds beautiful multicellular fruiting bodies that can be seen with the naked eye (click on my personal page underneath the page header for a publication list with pdf downloads, including a review paper). I thought it would be nice to return to the strains I isolated back then and do some population genomics. It turned out to be an interesting project although not without significant obstacles; papers are a bit like sausages, they can be very enjoyable but you do not want to necessarily know how they were made. Anyway, from the Abstract:
The bacterium Myxococcus xanthus glides through soil in search of prey microbes, but when food sources run out, cells cooperatively construct and sporulate within multicellular fruiting bodies. M. xanthus strains isolated from a 16×16 centimetre-scale patch of soil were previously shown to have diversified into many distinct compatibility types that are distinguished by the failure of swarming colonies to merge upon encounter. We sequenced the genomes of 22 isolates from this population belonging to the two most frequently occurring MultiLocus Sequence Type (MLST) clades in order to trace patterns of incipient genomic divergence, specifically related to social divergence. Although homologous recombination occurs frequently within the two MLST clades, we find an almost complete absence of recombination events between them. As the two clades are very closely related and live in sympatry, either ecological or genetic barriers must reduce genetic exchange between them. We find that the rate of change in the accessory genome is greater than the rate of amino acid substitution in the core genome. We identify a large genomic tract that consistently differs between isolates that do not freely merge and therefore is a candidate region for harbouring gene(s) responsible for self/non-self discrimination.
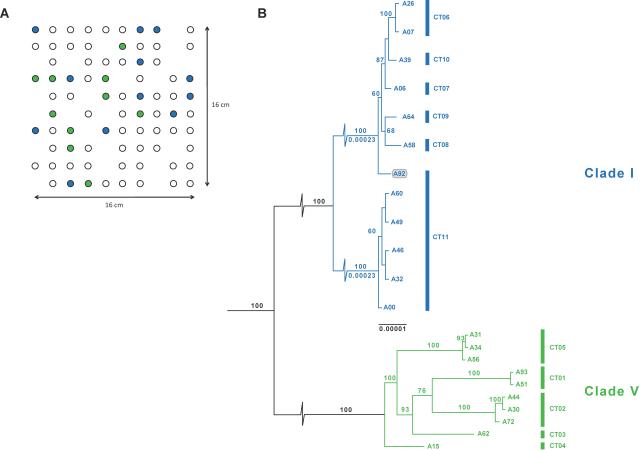 Above a nice Figure made by first author Seb Wielgoss that shows the location in the centimeter-scale plot the representatives of the two clades (genotypes) were isolated from (A) and a core genome phylogenetic tree clearly showing that the two clades are distinct, with limited within-clade diversity (B). For instance, strains A31 and A34 do not have a single SNP difference across their aligned conserved sequence of 7.6 Mb! For the homologous recombination analysis, co-author Xavier Didelot employed the FineStructure software (for a nice explanation on how this works see here), which has also been used in a high profile study on the genetic ancestry of the British population. The figure below shows a co-ancestry matrix on the left, with warmer colours representing higher percentages of recombinational copying from one genome to another. Genomes belonging to the same FineStructure population are connected by a vertical branch in the tree on the right, with the rest of the tree indicating inferred relationships between seven identified populations. The dark blue colours indicate negligible (<0.01%), homologous recombination between Clades I (populations 5-7) and V (populations 1-4). To my knowledge, this is only the third time a barrier to recombination between bacteria from the same population has been shown, and the first for a soil bacterium.
Above a nice Figure made by first author Seb Wielgoss that shows the location in the centimeter-scale plot the representatives of the two clades (genotypes) were isolated from (A) and a core genome phylogenetic tree clearly showing that the two clades are distinct, with limited within-clade diversity (B). For instance, strains A31 and A34 do not have a single SNP difference across their aligned conserved sequence of 7.6 Mb! For the homologous recombination analysis, co-author Xavier Didelot employed the FineStructure software (for a nice explanation on how this works see here), which has also been used in a high profile study on the genetic ancestry of the British population. The figure below shows a co-ancestry matrix on the left, with warmer colours representing higher percentages of recombinational copying from one genome to another. Genomes belonging to the same FineStructure population are connected by a vertical branch in the tree on the right, with the rest of the tree indicating inferred relationships between seven identified populations. The dark blue colours indicate negligible (<0.01%), homologous recombination between Clades I (populations 5-7) and V (populations 1-4). To my knowledge, this is only the third time a barrier to recombination between bacteria from the same population has been shown, and the first for a soil bacterium.
 Actually the main reason I wanted to sequence these genomes is to find out how different strains are able to recognize kin from non-kin. It is likely that Myxococcus meets other distinct strains, as many types can be found on a scale of even centimeters. In another paper, it could be shown that genetically distinct swarms were generally very bad at building fruiting bodies together. That scenario might actually not even be very relevant, as mixing of distinct cells is most likely when they are actively swarming. In the same paper however, it could be shown that swarms did not mix upon encounter, and that there thus must be some genetic mechanism whereby cells can discriminate self from non-self:
Actually the main reason I wanted to sequence these genomes is to find out how different strains are able to recognize kin from non-kin. It is likely that Myxococcus meets other distinct strains, as many types can be found on a scale of even centimeters. In another paper, it could be shown that genetically distinct swarms were generally very bad at building fruiting bodies together. That scenario might actually not even be very relevant, as mixing of distinct cells is most likely when they are actively swarming. In the same paper however, it could be shown that swarms did not mix upon encounter, and that there thus must be some genetic mechanism whereby cells can discriminate self from non-self:
By sequencing a set of genomes of strains that can either mix or not, genetic differences that consistently differ between such sets can be identified. Indeed, a large genomic tract was located that consistently differed with swarming compatibility. Again Seb made a really nice Figure to visualize these differences in gene content. First a phylogeny based on gene content variation in the variable region (A). Clusters match compatibility type (CT) groupings derived from social swarm merger assays. On the right a visual representation of the absence (black) and presence of single (light red) and multicopy (dark red) genes in the focal region. The proof in the pudding was a strain (A92) that could not mix with strains that it was otherwise genomically closely related to that was identical in this genomic tract with other strains it could mix with. This study was explorative in nature; you never know what you are going to find when sequencing new genomes. So there were a number of other significant findings (having multiple interesting findings perhaps paradoxically made the manuscript a harder sell…). For instance, we could show that changes in gene content (in the ‘accessory genome’) were much more prevalent than mutational changes (in the ‘core genome’). This finding actually was the inspiration of a recent Opinion paper on accessory genome evolution, see this recent post. I had hoped that differences on the DNA level would shed light on ecological differences between strains that could explain their coexistence but that proved very difficult as most genes were of unknown function. We did find massive differences in CRISPR-Cas systems though (these results ended up mainly in the supplemental material). The advance online publication is freely available from the journal’s website:
This study was explorative in nature; you never know what you are going to find when sequencing new genomes. So there were a number of other significant findings (having multiple interesting findings perhaps paradoxically made the manuscript a harder sell…). For instance, we could show that changes in gene content (in the ‘accessory genome’) were much more prevalent than mutational changes (in the ‘core genome’). This finding actually was the inspiration of a recent Opinion paper on accessory genome evolution, see this recent post. I had hoped that differences on the DNA level would shed light on ecological differences between strains that could explain their coexistence but that proved very difficult as most genes were of unknown function. We did find massive differences in CRISPR-Cas systems though (these results ended up mainly in the supplemental material). The advance online publication is freely available from the journal’s website:
Michiel
